
About me
Stephan Weißbach
PhD-student
s.weissbach@uni-mainz.de
GitHub | LinkedIn
room: 03.116, Biozentrum 1, Hanns-Dieter-Hüsch-Weg 15, 55128 Mainz
Current projects
- Premature upregulation of miR-92a’s target RBFOX2 hijacks PTBP splicing and impairs cortical neuronal differentiation
RBFOX2 is a RNA binding protein involved in neuronal differentiation and migration in the cerebral cortex. We use RNA-sequencing coupled with bioinformatic analysis to understand the role and partners of RBFOX2 during corticogenesis. - Cortexa – a comprehensive resource for studying gene expression and alternative splicing in the murine brain (www.cortexa-rna.com)
Cortexa is a comprehensive web-portal that incorporates RNA-sequencing datasets from the mouse cerebral cortex (longitudinal or cell-specific) and the hippocampus with a focus on alternative splicing analysis.
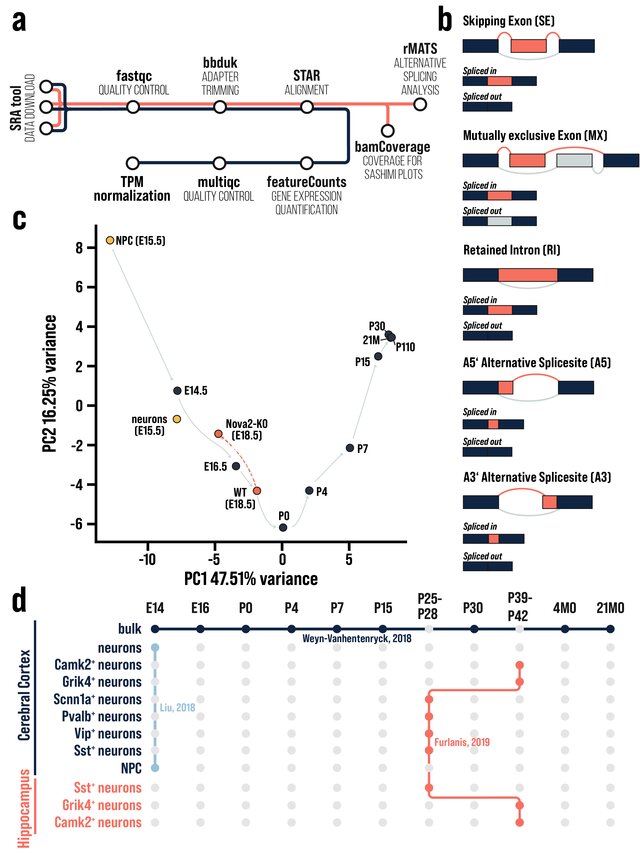
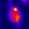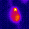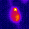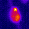
www.cortexa-rna.com - Neuroimaging Denoiser – Deep Learning aided denoising of iGlu-Snfr data

Noise is an abundant phenomenon impacting all experimental systems used in neuroscience. In imaging systems, noise affects pixels individually without any spatiotemporal relations. We employ U-Net to remove noise from microscopic imaging data.raw denoised 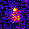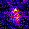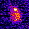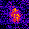
Publications
- Hristo Todorov, Stephan Weißbach, Laura Schlichtholz, Hanna Mueller, Dewi Hartwich, Susanne Gerber, Jennifer Winter: Stage-specific expression patterns and co-targeting relationships among miRNAs in the developing mouse cerebral cortex. Commun Biol 7, 1366 (2024). https://doi.org/10.1038/s42003-024-07092-7
- Stephan Weißbach, Hristo Todorov, Laura Schlichtholz, Sophia Mühlbauer, Lea Zografidou, Azza Soliman, Sarah Lor-Zade, Dewi Hartwich, Dennis Strand, Susanne Strand, Tanja Vogel, Martin Heine, Susanne Gerber, Jennifer Winter. Premature upregulation of miR-92a’s target RBFOX2 hijacks PTBP splicing and impairs cortical neuronal differentiation. bioRxiv 2024.09.20.614071; doi: https://doi.org/10.1101/2024.09.20.614071
- Neuroimage Denoiser for removing noise from transient fluorescent signals in functional imaging. Stephan Weißbach, Jonas Milkovits, Michela Borghi, Carolina Amaral, Abderazzaq El Khallouqi, Susanne Gerber, Martin Heine bioRxiv 2024.06.08.598061; doi: https://doi.org/10.1101/2024.06.08.598061
- Stanislav Jur 'Evic Sys, Alejandro Ceron-Noriega, Anne Kerber, Stephan Weissbach, Susann Schweiger, Michael Wand, Karin Everschor-Sitte and Susanne Gerber. Chromatin Capture Upsampling Toolbox - CCUT: A Versatile and unified Framework to Train Your Chromatin Capture Deep Learning Models. bioRxiv, https://doi.org/10.1101/2024.05.29.596528 (2024).
- Stephan Weißbach, Jonas Milkovits, Stefan Pastore, Martin Heine, Susanne Gerber, Hristo Todorov, Cortexa - a comprehensive resource for studying gene expression and alternative splicing in the murine brain. BMC Bioinformatics 25, 293 (2024). https://doi.org/10.1186/s12859-024-05919-y
- Nicolas Ruffini*, Saleh Altahini*, Stephan Weißbach*, Nico Weber, Jonas Milkovits, Anna Wierczeiko, Hendrik Backhaus, Albrecht Stroh, ViNe-Seg: Deep-Learning assisted segmentation of visible neurons and subsequent analysis embedded in a graphical user interface, Bioinformatics, 2024;, btae177, https://doi.org/10.1093/bioinformatics/btae177
- , , , , & (2022). CollembolAI, a macrophotography and computer vision workflow to digitize and characterize samples of soil invertebrate communities preserved in fluid. Methods in Ecology and Evolution, 00, 1– 14. https://doi.org/10.1111/2041-210X.14001
- Heck, J., Palmeira Do Amaral, A. C., Weißbach, S., El Khallouqi, A., Bikbaev, A., & Heine, M. (2021). More than a pore: How voltage-gated calcium channels act on different levels of neuronal communication regulation. Channels, 15(1), 322–338. https://doi.org/10.1080/19336950.2021.1900024
- Stephan Weißbach*, Stanislav Sys*, Charlotte Hewel, Hristo Todorov, Susann Schweiger, Jennifer Winter, Markus Pfenninger, Ali Torkamani, Doug Evans, Joachim Burger Karin Everschor-Sitte, Helen May-Simera and Susanne Gerber. Reliability of genomic variants across different next-generation sequencing platforms and bioinformatic processing pipelines. BMC Genomics 22, 62 (2021). https://doi.org/10.1186/s12864-020-07362-8
* equal contribution




